
Tidymodels Workflow with Functional Keras Models (Multi-Input)
workflows_functional.RmdIntroduction
This vignette demonstrates a complete tidymodels
workflow for a regression task using a Keras functional model defined
with kerasnip. We will use the Ames Housing dataset to
predict house prices. A key feature of this example is the use of a
multi-input Keras model, where numerical and categorical features are
processed through separate input branches.
kerasnip allows you to define complex Keras
architectures, including those with multiple inputs, and integrate them
seamlessly into the tidymodels ecosystem for robust
modeling and tuning.
Setup
First, we load the necessary packages.
library(kerasnip)
library(tidymodels)
#> ── Attaching packages ────────────────────────────────────── tidymodels 1.4.1 ──
#> ✔ broom 1.0.11 ✔ recipes 1.3.1
#> ✔ dials 1.4.2 ✔ rsample 1.3.1
#> ✔ dplyr 1.1.4 ✔ tailor 0.1.0
#> ✔ ggplot2 4.0.1 ✔ tidyr 1.3.1
#> ✔ infer 1.0.9 ✔ tune 2.0.1
#> ✔ modeldata 1.5.1 ✔ workflows 1.3.0
#> ✔ parsnip 1.4.0 ✔ workflowsets 1.1.1
#> ✔ purrr 1.2.0 ✔ yardstick 1.3.2
#> ── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
#> ✖ purrr::discard() masks scales::discard()
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ✖ recipes::step() masks stats::step()
library(keras3)
#>
#> Attaching package: 'keras3'
#> The following object is masked from 'package:yardstick':
#>
#> get_weights
library(dplyr) # For data manipulation
library(ggplot2) # For plotting
library(future) # For parallel processing
#>
#> Attaching package: 'future'
#> The following object is masked from 'package:keras3':
#>
#> %<-%
library(finetune) # For racingData Preparation
We’ll use the Ames Housing dataset, which is available in the
modeldata package. We will then split the data into
training and testing sets.
# Select relevant columns and remove rows with missing values
ames_df <- ames |>
select(
Sale_Price,
Gr_Liv_Area,
Year_Built,
Neighborhood,
Bldg_Type,
Overall_Cond,
Total_Bsmt_SF,
contains("SF")
) |>
na.omit()
# Split data into training and testing sets
set.seed(123)
ames_split <- initial_split(ames_df, prop = 0.8, strata = Sale_Price)
ames_train <- training(ames_split)
ames_test <- testing(ames_split)
# Create cross-validation folds for tuning
ames_folds <- vfold_cv(ames_train, v = 5, strata = Sale_Price)Recipe for Preprocessing
We will create a recipes object to preprocess our data.
This recipe will: * Predict Sale_Price using all other
variables. * Normalize all numerical predictors. * Create dummy
variables for categorical predictors. * Collapse each group of
predictors into a single matrix column using
step_collapse().
This final step is crucial for the multi-input Keras model, as the
kerasnip functional API expects a list of matrices for
multiple inputs, where each matrix corresponds to a distinct input
layer.
ames_recipe <- recipe(Sale_Price ~ ., data = ames_train) |>
step_normalize(all_numeric_predictors()) |>
step_collapse(all_numeric_predictors(), new_col = "numerical_input") |>
step_dummy(Neighborhood) |>
step_collapse(starts_with("Neighborhood"), new_col = "neighborhood_input") |>
step_dummy(Bldg_Type) |>
step_collapse(starts_with("Bldg_Type"), new_col = "bldg_input") |>
step_dummy(Overall_Cond) |>
step_collapse(starts_with("Overall_Cond"), new_col = "condition_input")Define Keras Functional Model with kerasnip
Now, we define our Keras functional model using
kerasnip’s layer blocks. This model will have four distinct
input layers: one for numerical features and three for categorical
features. These branches will be processed separately and then
concatenated before the final output layer.
# Define layer blocks for multi-input functional model
# Input blocks for numerical and categorical features
input_numerical <- function(input_shape) {
layer_input(shape = input_shape, name = "numerical_input")
}
input_neighborhood <- function(input_shape) {
layer_input(shape = input_shape, name = "neighborhood_input")
}
input_bldg <- function(input_shape) {
layer_input(shape = input_shape, name = "bldg_input")
}
input_condition <- function(input_shape) {
layer_input(shape = input_shape, name = "condition_input")
}
# Processing blocks for each input type
dense_numerical <- function(tensor, units = 32, activation = "relu") {
tensor |>
layer_dense(units = units, activation = activation)
}
dense_categorical <- function(tensor, units = 16, activation = "relu") {
tensor |>
layer_dense(units = units, activation = activation)
}
# Concatenation block
concatenate_features <- function(numeric, neighborhood, bldg, condition) {
layer_concatenate(list(numeric, neighborhood, bldg, condition))
}
# Output block for regression
output_regression <- function(tensor) {
layer_dense(tensor, units = 1, name = "output")
}
# Create the kerasnip model specification function
create_keras_functional_spec(
model_name = "ames_functional_mlp",
layer_blocks = list(
numerical_input = input_numerical,
neighborhood_input = input_neighborhood,
bldg_input = input_bldg,
condition_input = input_condition,
processed_numerical = inp_spec(dense_numerical, "numerical_input"),
processed_neighborhood = inp_spec(dense_categorical, "neighborhood_input"),
processed_bldg = inp_spec(dense_categorical, "bldg_input"),
processed_condition = inp_spec(dense_categorical, "condition_input"),
combined_features = inp_spec(
concatenate_features,
c(
numeric = "processed_numerical",
neighborhood = "processed_neighborhood",
bldg = "processed_bldg",
condition = "processed_condition"
)
),
output = inp_spec(output_regression, "combined_features")
),
mode = "regression"
)Model Specification
We’ll define our ames_functional_mlp model specification
and set some hyperparameters to tune(). Note how the
arguments are prefixed with their corresponding block names (e.g.,
processed_numerical_units).
# Define the tunable model specification
functional_mlp_spec <- ames_functional_mlp(
# Tunable parameters for numerical branch
processed_numerical_units = tune(),
# Tunable parameters for categorical branch
processed_neighborhood_units = tune(),
processed_bldg_units = tune(),
processed_condition_units = tune(),
# Fixed compilation and fitting parameters
compile_loss = "mean_squared_error",
compile_optimizer = "adam",
compile_metrics = c("mean_absolute_error"),
fit_epochs = 50,
fit_batch_size = 32,
fit_validation_split = 0.2,
fit_callbacks = list(
callback_early_stopping(monitor = "val_loss", patience = 5)
)
) |>
set_engine("keras")
print(functional_mlp_spec)
#> ames functional mlp Model Specification (regression)
#>
#> Main Arguments:
#> num_numerical_input = structure(list(), class = "rlang_zap")
#> num_neighborhood_input = structure(list(), class = "rlang_zap")
#> num_bldg_input = structure(list(), class = "rlang_zap")
#> num_condition_input = structure(list(), class = "rlang_zap")
#> num_processed_numerical = structure(list(), class = "rlang_zap")
#> num_processed_neighborhood = structure(list(), class = "rlang_zap")
#> num_processed_bldg = structure(list(), class = "rlang_zap")
#> num_processed_condition = structure(list(), class = "rlang_zap")
#> num_combined_features = structure(list(), class = "rlang_zap")
#> num_output = structure(list(), class = "rlang_zap")
#> processed_numerical_units = tune()
#> processed_numerical_activation = structure(list(), class = "rlang_zap")
#> processed_neighborhood_units = tune()
#> processed_neighborhood_activation = structure(list(), class = "rlang_zap")
#> processed_bldg_units = tune()
#> processed_bldg_activation = structure(list(), class = "rlang_zap")
#> processed_condition_units = tune()
#> processed_condition_activation = structure(list(), class = "rlang_zap")
#> learn_rate = structure(list(), class = "rlang_zap")
#> fit_batch_size = 32
#> fit_epochs = 50
#> fit_callbacks = list(callback_early_stopping(monitor = "val_loss", patience = 5))
#> fit_validation_split = 0.2
#> fit_validation_data = structure(list(), class = "rlang_zap")
#> fit_shuffle = structure(list(), class = "rlang_zap")
#> fit_class_weight = structure(list(), class = "rlang_zap")
#> fit_sample_weight = structure(list(), class = "rlang_zap")
#> fit_initial_epoch = structure(list(), class = "rlang_zap")
#> fit_steps_per_epoch = structure(list(), class = "rlang_zap")
#> fit_validation_steps = structure(list(), class = "rlang_zap")
#> fit_validation_batch_size = structure(list(), class = "rlang_zap")
#> fit_validation_freq = structure(list(), class = "rlang_zap")
#> fit_verbose = structure(list(), class = "rlang_zap")
#> fit_view_metrics = structure(list(), class = "rlang_zap")
#> compile_optimizer = adam
#> compile_loss = mean_squared_error
#> compile_metrics = c("mean_absolute_error")
#> compile_loss_weights = structure(list(), class = "rlang_zap")
#> compile_weighted_metrics = structure(list(), class = "rlang_zap")
#> compile_run_eagerly = structure(list(), class = "rlang_zap")
#> compile_steps_per_execution = structure(list(), class = "rlang_zap")
#> compile_jit_compile = structure(list(), class = "rlang_zap")
#> compile_auto_scale_loss = structure(list(), class = "rlang_zap")
#>
#> Computational engine: kerasCreate Workflow
A workflow combines the recipe and the model
specification.
ames_wf <- workflow() |>
add_recipe(ames_recipe) |>
add_model(functional_mlp_spec)
print(ames_wf)
#> ══ Workflow ════════════════════════════════════════════════════════════════════
#> Preprocessor: Recipe
#> Model: ames_functional_mlp()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> 8 Recipe Steps
#>
#> • step_normalize()
#> • step_collapse()
#> • step_dummy()
#> • step_collapse()
#> • step_dummy()
#> • step_collapse()
#> • step_dummy()
#> • step_collapse()
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> ames functional mlp Model Specification (regression)
#>
#> Main Arguments:
#> num_numerical_input = structure(list(), class = "rlang_zap")
#> num_neighborhood_input = structure(list(), class = "rlang_zap")
#> num_bldg_input = structure(list(), class = "rlang_zap")
#> num_condition_input = structure(list(), class = "rlang_zap")
#> num_processed_numerical = structure(list(), class = "rlang_zap")
#> num_processed_neighborhood = structure(list(), class = "rlang_zap")
#> num_processed_bldg = structure(list(), class = "rlang_zap")
#> num_processed_condition = structure(list(), class = "rlang_zap")
#> num_combined_features = structure(list(), class = "rlang_zap")
#> num_output = structure(list(), class = "rlang_zap")
#> processed_numerical_units = tune()
#> processed_numerical_activation = structure(list(), class = "rlang_zap")
#> processed_neighborhood_units = tune()
#> processed_neighborhood_activation = structure(list(), class = "rlang_zap")
#> processed_bldg_units = tune()
#> processed_bldg_activation = structure(list(), class = "rlang_zap")
#> processed_condition_units = tune()
#> processed_condition_activation = structure(list(), class = "rlang_zap")
#> learn_rate = structure(list(), class = "rlang_zap")
#> fit_batch_size = 32
#> fit_epochs = 50
#> fit_callbacks = list(callback_early_stopping(monitor = "val_loss", patience = 5))
#> fit_validation_split = 0.2
#> fit_validation_data = structure(list(), class = "rlang_zap")
#> fit_shuffle = structure(list(), class = "rlang_zap")
#> fit_class_weight = structure(list(), class = "rlang_zap")
#> fit_sample_weight = structure(list(), class = "rlang_zap")
#> fit_initial_epoch = structure(list(), class = "rlang_zap")
#> fit_steps_per_epoch = structure(list(), class = "rlang_zap")
#> fit_validation_steps = structure(list(), class = "rlang_zap")
#> fit_validation_batch_size = structure(list(), class = "rlang_zap")
#> fit_validation_freq = structure(list(), class = "rlang_zap")
#> fit_verbose = structure(list(), class = "rlang_zap")
#> fit_view_metrics = structure(list(), class = "rlang_zap")
#> compile_optimizer = adam
#> compile_loss = mean_squared_error
#> compile_metrics = c("mean_absolute_error")
#> compile_loss_weights = structure(list(), class = "rlang_zap")
#> compile_weighted_metrics = structure(list(), class = "rlang_zap")
#> compile_run_eagerly = structure(list(), class = "rlang_zap")
#> compile_steps_per_execution = structure(list(), class = "rlang_zap")
#> compile_jit_compile = structure(list(), class = "rlang_zap")
#> compile_auto_scale_loss = structure(list(), class = "rlang_zap")
#>
#> Computational engine: kerasDefine Tuning Grid
We will create a regular grid for our hyperparameters.
# Define the tuning grid
params <- extract_parameter_set_dials(ames_wf) |>
update(
processed_numerical_units = hidden_units(range = c(32, 128)),
processed_neighborhood_units = hidden_units(range = c(16, 64)),
processed_bldg_units = hidden_units(range = c(16, 64)),
processed_condition_units = hidden_units(range = c(16, 64))
)
functional_mlp_grid <- grid_regular(params, levels = 3)
print(functional_mlp_grid)
#> # A tibble: 81 × 4
#> processed_numerical_units processed_neighborhood_units processed_bldg_units
#> <int> <int> <int>
#> 1 32 16 16
#> 2 80 16 16
#> 3 128 16 16
#> 4 32 40 16
#> 5 80 40 16
#> 6 128 40 16
#> 7 32 64 16
#> 8 80 64 16
#> 9 128 64 16
#> 10 32 16 40
#> # ℹ 71 more rows
#> # ℹ 1 more variable: processed_condition_units <int>Tune Model
Now, we’ll use tune_race_anova() to perform
cross-validation and find the best hyperparameters.
# Note: Parallel processing with `plan(multisession)` is currently not working
# with Keras models due to backend conflicts
set.seed(123)
ames_tune_results <- tune_race_anova(
ames_wf,
resamples = ames_folds,
grid = functional_mlp_grid,
metrics = metric_set(rmse, mae, rsq),
control = control_race(save_pred = TRUE, save_workflow = TRUE)
)
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 7ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/step
#> 15/15 - 0s - 6ms/stepInspect Tuning Results
We can inspect the tuning results to see which hyperparameter combinations performed best.
# Show the best performing models based on RMSE
show_best(ames_tune_results, metric = "rmse", n = 5)
#> # A tibble: 2 × 10
#> processed_numerical_units processed_neighborhood_units processed_bldg_units
#> <int> <int> <int>
#> 1 128 64 64
#> 2 128 64 40
#> # ℹ 7 more variables: processed_condition_units <int>, .metric <chr>,
#> # .estimator <chr>, mean <dbl>, n <int>, std_err <dbl>, .config <chr>
# Autoplot the results
# Currently does not work due to a label issue: autoplot(ames_tune_results)
# Select the best hyperparameters
best_functional_mlp_params <- select_best(ames_tune_results, metric = "rmse")
print(best_functional_mlp_params)
#> # A tibble: 1 × 5
#> processed_numerical_units processed_neighborhood_units processed_bldg_units
#> <int> <int> <int>
#> 1 128 64 64
#> # ℹ 2 more variables: processed_condition_units <int>, .config <chr>Finalize Workflow and Fit Model
Once we have the best hyperparameters, we finalize the workflow and fit the model on the entire training dataset.
# Finalize the workflow with the best hyperparameters
final_ames_wf <- finalize_workflow(ames_wf, best_functional_mlp_params)
# Fit the final model on the full training data
final_ames_fit <- fit(final_ames_wf, data = ames_train)
print(final_ames_fit)
#> ══ Workflow [trained] ══════════════════════════════════════════════════════════
#> Preprocessor: Recipe
#> Model: ames_functional_mlp()
#>
#> ── Preprocessor ────────────────────────────────────────────────────────────────
#> 8 Recipe Steps
#>
#> • step_normalize()
#> • step_collapse()
#> • step_dummy()
#> • step_collapse()
#> • step_dummy()
#> • step_collapse()
#> • step_dummy()
#> • step_collapse()
#>
#> ── Model ───────────────────────────────────────────────────────────────────────
#> $fit
#> Model: "functional_257"
#> ┏━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┓
#> ┃ Layer (type) ┃ Output Shape ┃ Param # ┃ Connected to ┃
#> ┡━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━┩
#> │ numerical_input │ (None, 1, 10) │ 0 │ - │
#> │ (InputLayer) │ │ │ │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ neighborhood_input │ (None, 1, 28) │ 0 │ - │
#> │ (InputLayer) │ │ │ │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ bldg_input │ (None, 1, 4) │ 0 │ - │
#> │ (InputLayer) │ │ │ │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ condition_input │ (None, 1, 9) │ 0 │ - │
#> │ (InputLayer) │ │ │ │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ dense_1028 (Dense) │ (None, 1, 128) │ 1,408 │ numerical_input[0… │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ dense_1029 (Dense) │ (None, 1, 64) │ 1,856 │ neighborhood_inpu… │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ dense_1030 (Dense) │ (None, 1, 64) │ 320 │ bldg_input[0][0] │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ dense_1031 (Dense) │ (None, 1, 64) │ 640 │ condition_input[0… │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ concatenate_257 │ (None, 1, 320) │ 0 │ dense_1028[0][0], │
#> │ (Concatenate) │ │ │ dense_1029[0][0], │
#> │ │ │ │ dense_1030[0][0], │
#> │ │ │ │ dense_1031[0][0] │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ output (Dense) │ (None, 1, 1) │ 321 │ concatenate_257[0… │
#> └───────────────────────┴───────────────────┴─────────────┴────────────────────┘
#> Total params: 13,637 (53.27 KB)
#> Trainable params: 4,545 (17.75 KB)
#> Non-trainable params: 0 (0.00 B)
#> Optimizer params: 9,092 (35.52 KB)
#>
#> $history
#>
#> Final epoch (plot to see history):
#> loss: 1,183,665,664
#> mean_absolute_error: 21,118
#> val_loss: 6,149,310,464
#> val_mean_absolute_error: 59,552
#>
#> $lvl
#> NULL
#>
#> $process_x
#> function (x)
#>
#> ...
#> and 88 more lines.Inspect Final Model
You can extract the underlying Keras model and its training history for further inspection.
# Extract the Keras model summary
final_ames_fit |>
extract_fit_parsnip() |>
extract_keras_model() |>
summary()
#> Model: "functional_257"
#> ┏━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━┓
#> ┃ Layer (type) ┃ Output Shape ┃ Param # ┃ Connected to ┃
#> ┡━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━┩
#> │ numerical_input │ (None, 1, 10) │ 0 │ - │
#> │ (InputLayer) │ │ │ │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ neighborhood_input │ (None, 1, 28) │ 0 │ - │
#> │ (InputLayer) │ │ │ │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ bldg_input │ (None, 1, 4) │ 0 │ - │
#> │ (InputLayer) │ │ │ │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ condition_input │ (None, 1, 9) │ 0 │ - │
#> │ (InputLayer) │ │ │ │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ dense_1028 (Dense) │ (None, 1, 128) │ 1,408 │ numerical_input[0… │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ dense_1029 (Dense) │ (None, 1, 64) │ 1,856 │ neighborhood_inpu… │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ dense_1030 (Dense) │ (None, 1, 64) │ 320 │ bldg_input[0][0] │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ dense_1031 (Dense) │ (None, 1, 64) │ 640 │ condition_input[0… │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ concatenate_257 │ (None, 1, 320) │ 0 │ dense_1028[0][0], │
#> │ (Concatenate) │ │ │ dense_1029[0][0], │
#> │ │ │ │ dense_1030[0][0], │
#> │ │ │ │ dense_1031[0][0] │
#> ├───────────────────────┼───────────────────┼─────────────┼────────────────────┤
#> │ output (Dense) │ (None, 1, 1) │ 321 │ concatenate_257[0… │
#> └───────────────────────┴───────────────────┴─────────────┴────────────────────┘
#> Total params: 13,637 (53.27 KB)
#> Trainable params: 4,545 (17.75 KB)
#> Non-trainable params: 0 (0.00 B)
#> Optimizer params: 9,092 (35.52 KB)
# Plot the Keras model
final_ames_fit |>
extract_fit_parsnip() |>
extract_keras_model() |>
plot(show_shapes = TRUE)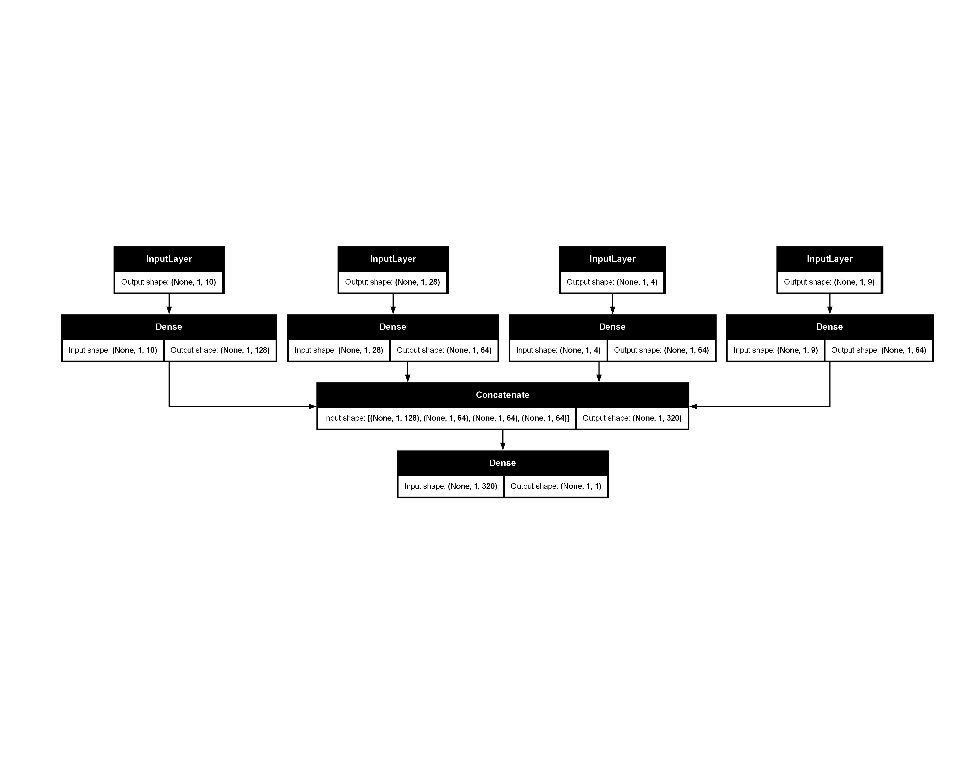
# Plot the training history
final_ames_fit |>
extract_fit_parsnip() |>
extract_keras_history() |>
plot()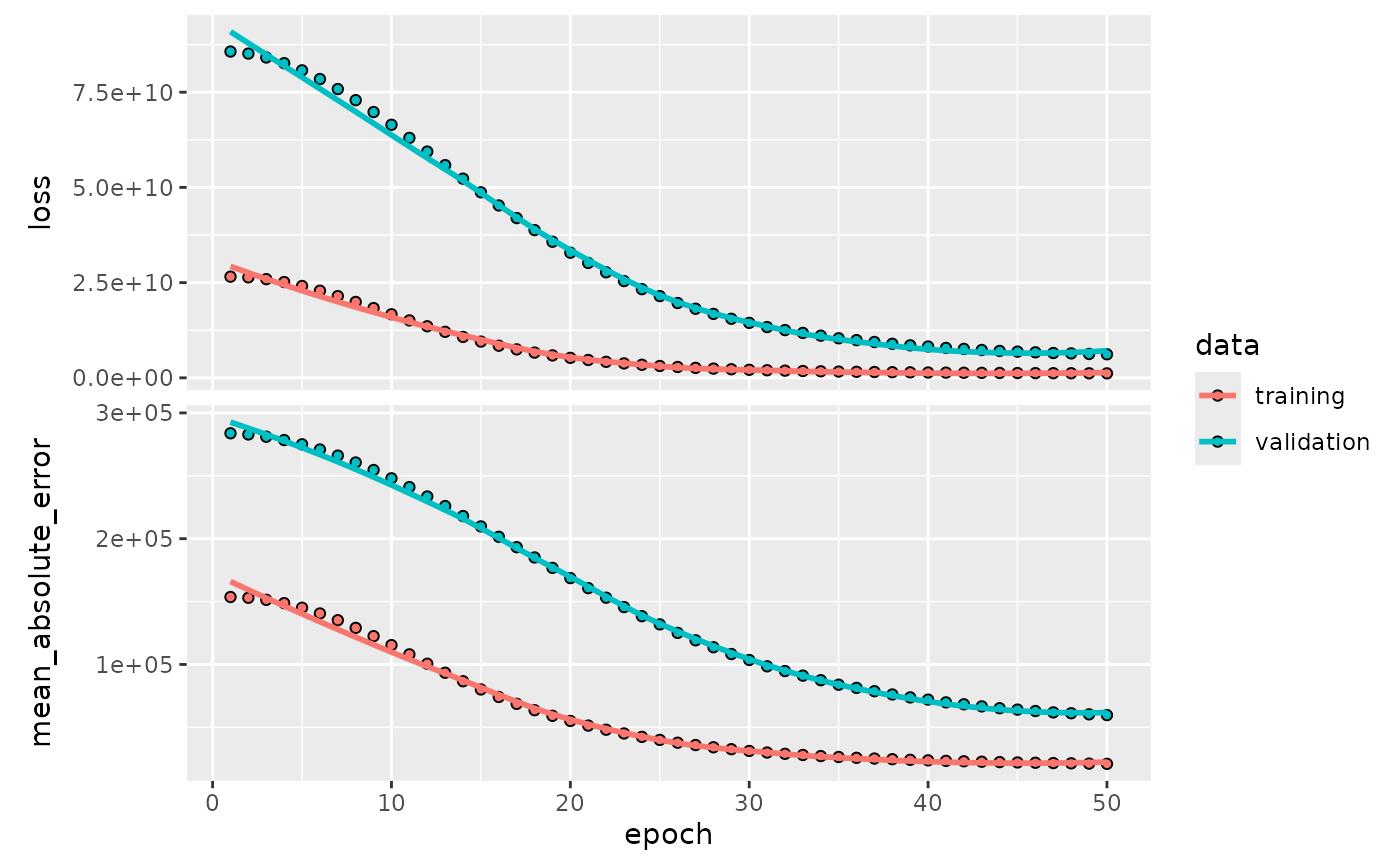
Make Predictions and Evaluate
Finally, we will make predictions on the test set and evaluate the model’s performance.
# Make predictions on the test set
ames_test_pred <- predict(final_ames_fit, new_data = ames_test)
#> 19/19 - 0s - 5ms/step
# Combine predictions with actuals
ames_results <- tibble::tibble(
Sale_Price = ames_test$Sale_Price,
.pred = ames_test_pred$.pred
)
print(head(ames_results))
#> # A tibble: 6 × 2
#> Sale_Price .pred
#> <int> <dbl>
#> 1 105000 98008.
#> 2 172000 161664.
#> 3 189900 193386.
#> 4 115000 127545.
#> 5 395192 267440.
#> 6 214000 213320.
# Evaluate performance using yardstick metrics
metrics_results <- metric_set(
rmse,
mae,
rsq
)(
ames_results,
truth = Sale_Price,
estimate = .pred
)
print(metrics_results)
#> # A tibble: 3 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 rmse standard 50750.
#> 2 mae standard 30780.
#> 3 rsq standard 0.787